Pathway enrichment analysis / visualize for Single Cell Data
Online manual is available in here
🔧 Install
devtools::install_github('noobCoding/cellenrich') library(CellEnrich)
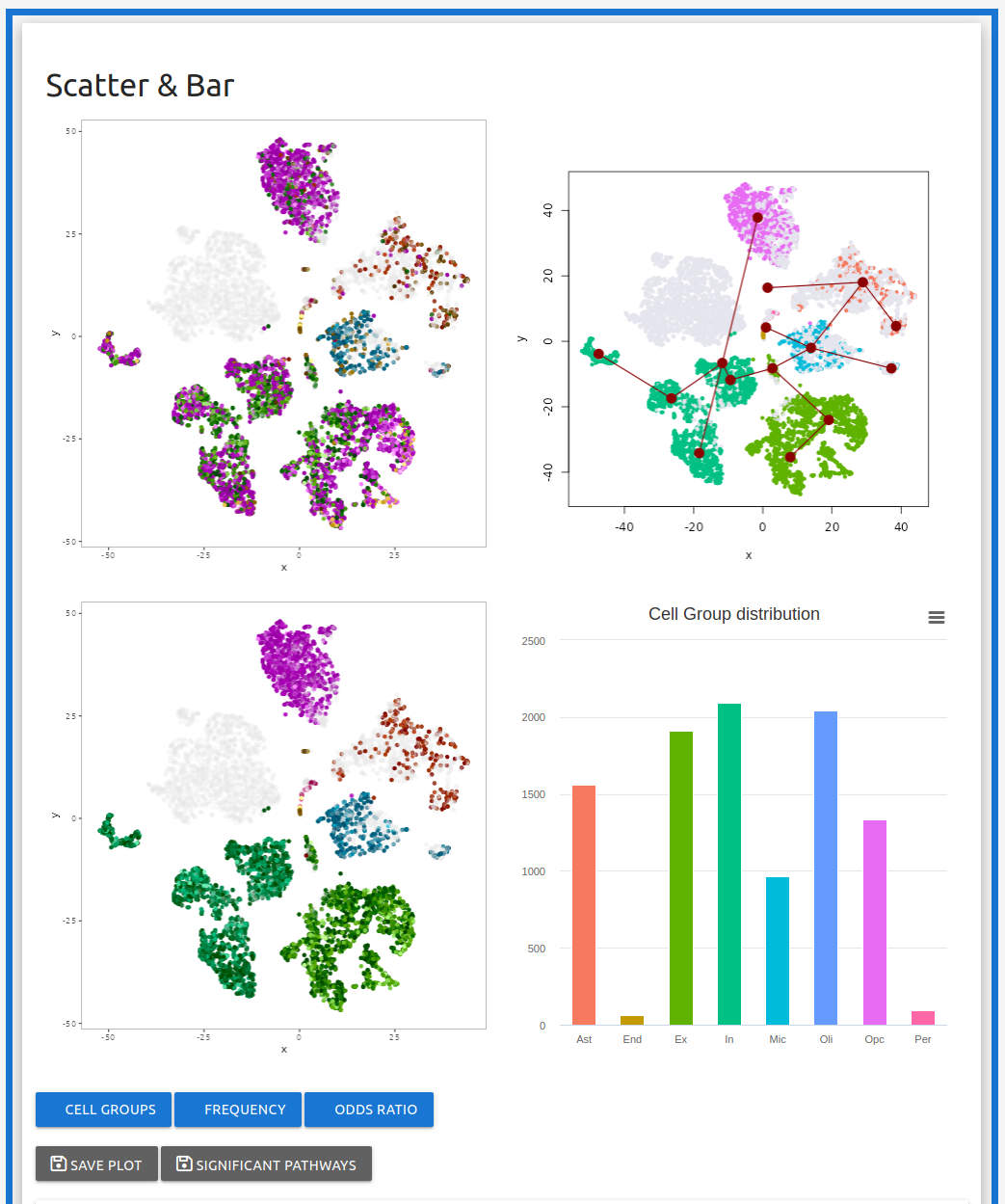
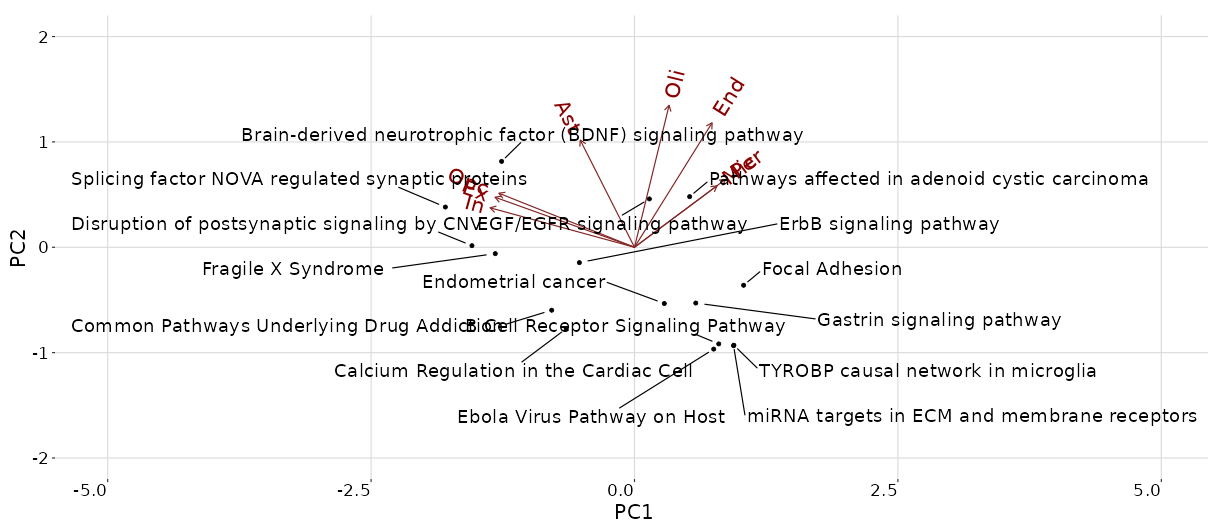
Example run with Alzheimer data
# download minimal data to working directory
download.file('https://github.com/noobcoding/CellEnrich/raw/master/data/Alzheimer_Counts_sampled.RDS','Alzheimer_Counts_sampled.RDS', mode = 'wb')
download.file('https://github.com/noobcoding/CellEnrich/raw/master/data/Alzheimer_CellType_sampled.RDS','Alzheimer_CellType_sampled.RDS', mode = 'wb')
download.file('https://github.com/noobcoding/CellEnrich/raw/master/data/Reactome_2022.RData', 'Reactome_2022.RData', mode = 'wb')
# load library and data
library(CellEnrich)
GroupInfo <- readRDS("Alzheimer_CellType_sampled.RDS")
CountData <- readRDS("Alzheimer_Counts_sampled.RDS")
# Run cellenrich
CellEnrich(CountData, GroupInfo)📝 License
This project is MIT licensed